Mapping the favourable character of intermolecular contacts
- Christian Jelsch
- Session 5 , Chemical Bonding
- Tuesday, July 15, 2025
- 9:30 am
Christian Jelscha, Benoît Guillota, Haritha Mambattaa, Maria Mitevab
aCRM2, CNRS, Lorraine University, 54000 Nancy. France; bCiTCoM, Université Paris Cité, CNRS Inserm, Paris, 75006, France
e-mail: christian.jelsch@univ-lorraine.fr
Structure-based methods including docking and scoring of ligands aim to select the best potential hits for in vitro assays by ranking them according to predicted binding affinities. Virtual screening is based on docking programs providing several poses of the ligand within a binding site and a score attributed to each pose. Accurate prediction of the correct binding pose remains a major scientific challenge for current scoring functions in order to retrieve the true ligands.
The newly developed descriptor [1] concerns a computer based method for evaluating the stability of a complex formed by two molecular entities. It provides a score and at the same time a fingerprint of the intermolecular interactions. The descriptor can be applied for instance to interacting molecules in crystals or to protein / ligand complexes, either experimental (native) or produced by docking software. The function is projected on the intermolecular Hirshfeld interface and is homogenous to an electrostatic energy density. This electrostatic complementarity descriptor can act as filter to detect most false ligand poses among diverse poses proposed by molecular docking programs. The crystal structure of the cytochrome P450 2C9 / warfarin complex [2] is analyzed in the Figure 1. A good electrostatic complementarity is typically manifested by a correlation coefficient between interior and exterior properties close to -1 and by a fingerprint plot with points located close to the diagonal line y=-x. The lines of points correspond to strong interactions.
Moreover, an empirical van der Waals (vdW) energy function has been developed in MoProSuite to complement the electrostatic and polarization energy. The parameter optimization for vdW interactions was conducted using a least-squares algorithm implemented in the SciPy python library. This approach minimizes the sum of squared residuals between the SAPT+2 vdW interaction energies from the NENCI-2021 dataset [5] and the corresponding vdW values computed by the fitting program.
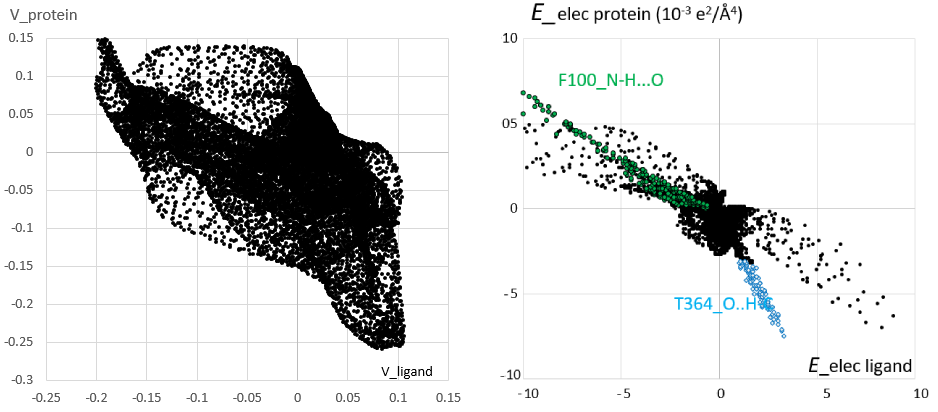
Figure 1. Left: scatterplot of electrostatic potential generated by the ligand warfarin and the protein CYP 2C9 on the Hirshfeld interface. Right: scatterplot of electrostatic energy density. The charge density was transferred from ELMAM2 database [3] of multipolar atoms. Electrostatic and Hirshfeld surface calculations were done with VMoPro and MoProViewer [4] software.
References:
[1] Jelsch, Miteva, Guillot (2023) European Patent EP004195210A1.
[2] Williams … Jhoti. 2003, Nature, 424, 464-468.
[3] Domagala, … Guillot, Jelsch. 2012. Acta Cryst. A68,337-351.
[4] Vukovic, … Guillot, Jelsch. 2021. Acta Cryst. D77, 1292-1304.
[5] Sparrow … DiStasio. 2021. J. Chemical Physics, 155(18), 184303.