Defining Tools for Complex Small Molecules to Enable Assessment of Quality and Appropriate Reuse of Data
- Aaron Horner
- Talk
- May 16, 2025
Aaron Horner
University of Southampton
e-mail: ah5g17@soton.ac.uk
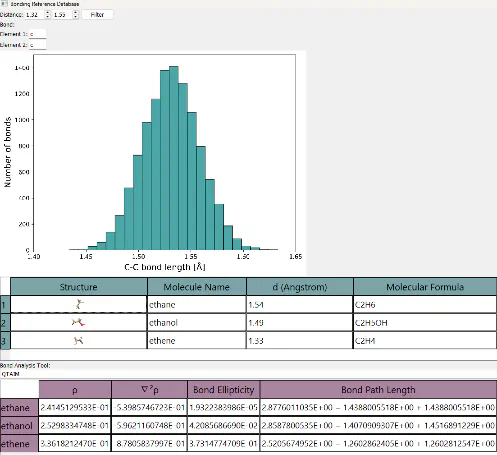
An important, but often difficult to evaluate, aspect of small molecule crystallographic structure analysis is an understanding of how a refinement has been performed, as the final result (CIF) contains little procedural information.
For more complicated analysis, such as dynamic crystallography or the crystal sponge technique, this understanding is key as the structures are often complex and may require a great number of restraints and constraints to obtain a result. The implementation of structure grading systems that can account for the refinement process, alongside the use of conventional quality metrics, would enable appropriate evaluation and verification of results which arise from these more complex experiments.
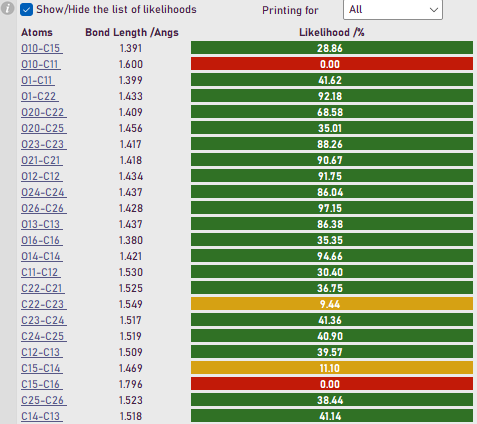
The output from a prototype grading tool that exists as a plugin within Olex2 1.5-alpha which evaluates the likelihood that a bond is ‘appropriate’ based on bonding data acquired from existing structural databases.
A secondary issue is that the number of structures which are published each year are ever increasing[1] and with the advent of AI and Machine Learning in crystallography and the generation of predictive crystal structures[2], it is imperative that we have tools which can aid analysts when refining crystal structures but also work as a CheckCIF style approach to evaluate and verify a structure before it is published.
One of the tools I wish to discuss during this meeting can be used to evaluate the likelihood of a given bond in a structure, and from this calculate the total likelihood of the structure, Figure 1. I believe it is important that we consider the tools we can implement to alleviate the aforementioned issues and provide tools to help more complicated analysis.
References:
[1] Ferrence, Gregory M et al. “CSD Communications of the Cambridge Structural Database.” IUCrJ 10, 6-15 (2023).
[2] Szymanski, N.J., Rendy, B., Fei, Y. et al. An autonomous laboratory for the accelerated synthesis of novel materials. Nature 624, 86–91 (2023).